Estimated Breeding Values (EBVs)
and the Cavalier King Charles Spaniel
-
 What
are EBVs?
What
are EBVs? - Importance of objective data
- Can a dog's EBV change?
- How are EBVs to be used?
- Advantages of using EBVs
- Genetics of SM -- By Dr. Sarah Blott
- Research News
- Related Links
- Veterinary Resources
The cavalier King Charles spaniel (CKCS) breed is susceptible to 25 inherited disorders. (See this February 2015 article and this December 2009 article.)
What are EBVs?
An “estimated breeding value” (EBV) is a statistical numerical prediction of the relative genetic value of a particular dog (male or female) available for breeding. EBVs are used to rank breeding stock for selection, based upon the genetic risk of each dog with regard to one or more specified traits. An EBV therefore is a calculated estimate of heritability for each trait being considered, relative to the EBVs of other dogs in the breeding population. EBVs are limited to the genetic liabilities and do not include environmental conditions subsequent to birth.
EBVs for cavalier King Charles spaniels are intended to be each dog’s genetic risk of disease – initially syringomyelia and early-onset mitral valve disease – relative to the rest of the available breeding stock.
EBVs are calculated by compiling as much objective information about each dog as possible, including health test data, together with its pedigree information and physical characteristics. The sum of this data is called the dog’s “phenotype” – what can be observed and measured with respect to the disease(s). A dog’s phenotype will consist of both genetic and non-genetic components.
The importance of using objective data
To be useful to breeders, EBVs must be accurate and unbiased. This accuracy applies to both the particular dog being examined as well as the breeding population as a whole. For EBVs to be accurate, the calculators constantly need as much information, particularly health test data, on as many cavaliers as possible.
Can a dog’s EBV Change?
EBVs are based upon the data available for estimating them. As new information is collected, about the particular dog or its relatives, its EBV can be re-calculated to improve the accuracy of its estimate. Initially, an off-spring’s EBV is the sum of its sire’s and dam’s scores, divided by two. As more and more of the dog’s health test data and other information is acquired, the EBV will change.
How are EBVs to be used?
The EBV system is evolving and currently in its early stage,
and presently only among cavaliers in the United Kingdom.
 The
UK Kennel Club has introduced its EBV database program, called
Mate Select, which is designed to generate EBVs. Mate Select is intended to enable breeders to assess the impact that a proposed mating will have on
the genetic diversity of the offspring and maximize the chances of producing
healthy puppies while also having the optimum impact on the breed’s genetic
diversity. In the UK, once EBVs are assigned , breeders can compare the
EBVs of the breeding stock, to determine which cavaliers may be expected to best
meet the SM and MVD breeding protocols when bred.
The
UK Kennel Club has introduced its EBV database program, called
Mate Select, which is designed to generate EBVs. Mate Select is intended to enable breeders to assess the impact that a proposed mating will have on
the genetic diversity of the offspring and maximize the chances of producing
healthy puppies while also having the optimum impact on the breed’s genetic
diversity. In the UK, once EBVs are assigned , breeders can compare the
EBVs of the breeding stock, to determine which cavaliers may be expected to best
meet the SM and MVD breeding protocols when bred.
EBVs will be an important, objective (and hopefully unbiased) tool for breeders to use in the process of selecting breeding stock. Their aim is to improve the accuracy of breeding stock selection decisions, resulting in reducing the risk of genetic disease in each succeeding generation. Sarah C. Blott, a geneticist at the UK's Animal Health Trust, said: “Science will completely transform dog breeding in the future, allowing breeders to make choices about which dogs to mate based on scientific information that will be available from the Kennel Club.”
Of course, it will remain the decision of each breeder as to whether to use EBVs and Mate Select. Hopefully, more breeders will take advantage of this information than have been following the currently available breeding guidelines, such as the mitral valve disease breeding protocol and the syringomyelia breeding protocol, both of which have been largely ignored, at least in the United States.
The advantages of using EBVs
 Dr. Clare Rusbridge, UK veterinary neurologist has compared
the current SM breeding protocol to EBVs for
combating syringomyelia on
her blog in 2011:
Dr. Clare Rusbridge, UK veterinary neurologist has compared
the current SM breeding protocol to EBVs for
combating syringomyelia on
her blog in 2011:
"It has been five years since the introduction of the International Breeding guidelines for Syringomyelia in 2006. These provided an interim measure for breeders based on existing evidence with a common sense approach until there was better understanding for this complex condition. A Mate Select Scheme that generates estimated breeding values is considered superior. Some breeders consider themselves capable of making their own decisions based on their wealth of breeding experience and knowledge of their own dogs’ MRIs. However CMSM is a complex trait which means that knowing the health status of close relatives is unlikely to be sufficient for predicting the disease free status of the offspring. Information about generations of dogs needs to be taken into account."
In a 2010 article about evaluating hip scores of Labrador retrievers, the UK authors, Thomas W. Lewis, Sarah C. Blott, and John A. Woolliams, explained the advantages of using EBVs for selecting breeding stock:
"The presented results have demonstrated that the availability of EBV through routine evaluations of the hip score data would hasten progress in alleviating the problem of hip dysplasia via increases in selective accuracy compared to selection based on phenotype alone [for example, MRI scans or heart testing]. However the benefits of EBV extend beyond the simple comparisons of accuracy for a recently scored dog:
(i) the EBV for an individual, unlike its phenotypic score, will further increase in accuracy over time by utilising all the available information and being updated as additional information becomes available e.g. from offspring or siblings;
(ii) the EBV will provide predictors for those animals that do not have a phenotypic record hence increasing selection opportunities and intensity, which again enhances rate of improvement;
(iii) the EBV will be available from the moment of birth for selection (although newborn littermates will have identical EBV) and, in this case, the accuracy (and hence rate of improvement) from using EBV increases by 31% compared to the parental average phenotype;
(iv) the EBV will have been corrected for other fixed effects such as sex and age which bias phenotype as a predictor of genetic merit; and
(v) it may be argued that taking account of a sustainable rate of inbreeding as well as disease prevalence would restrict the selection pressure that can be applied, however this only serves to place a greater emphasis on the accuracy of the selection that does take place.
"Finally with the availability of sequence and dense canine SNP chips, the development of a genomic EBV (an EBV informed by additional information from dense SNP genotypes) would help to distinguish littermates and further increase accuracy, increasing the potential rate of improvement, and might also lead subsequently to scientific benefits through identifying the major QTL. The intention is to make public the EBV for hip score for all KC registered Labrador Retrievers so that all these benefits may be realised."
RETURN TO TOP
Genetics of SM
Genetics of Syringomyelia and Breeding Strategies
to Reduce Occurrence
By SARAH BLOTT, PhD (Quantitative Genetics), MSc (Animal
Breeding)
Genetics Department, Animal Health Trust, UK
Notes for the [UK] Cavalier King Charles Spaniel Clubs Liaison Meeting 18 May 2008
Syringomyelia is believed to be a complex disease, where the disease phenotype results from the effects of several genes plus environmental influences. The phenotype includes not only the affectation status of the individual but also clinical observations and measurements made from MRI scans. In order to determine the genetic basis of the disease two different approaches are being taken. The first uses a population-based approach, where phenotypic measurements and pedigree information are used to estimate the heritability of the disease. This requires that we have accurate phenotypic measurements, including MRI scans, on as much of the population as possible, together with pedigrees so that genetic relationships between individuals can be identified.
Where information exists on other diseases, such as Mitral Valve Disease, this can also be included in the analysis allowing genetic correlations between diseases to be established. It is important to know about correlations, or relationships, between diseases so that any selection strategies take account of the possible influence that selection against one disease may have on other diseases. The second approach to understanding the genetic basis of a disease is to use molecular genetics and gene mapping techniques to try to identify the underlying causative mutations. This approach is also being used to try to identify genes causing both Syringomyelia (SM) and Chiari Malformation (CM) in the Cavalier King Charles Spaniel (CKCS) and other toy breeds such as the Brussels Griffon. It is hoped that this will identify regions of the genome harbouring the genes causing these conditions.
Data collected by Penny Knowler and Clare Rusbridge is currently being used as the basis for the population-based analysis of heritability. Their database contains clinical observations for SM and CM on around 1,400 dogs and MRI scan results for around 700 of these dogs. We have also been given access to the full UK Kennel Club pedigree records for CKCS. This enables us to estimate the heritability of SM and the genetic correlations between SM and measurements made from the MRI scans. The information obtained from this analysis then allows us to derive estimated breeding values (EBVs) for all measured dogs as well as all dogs in the pedigree.
Once the results of the gene mapping studies become available it is hoped to bring this information together with the population analysis to facilitate the calculation of genomic breeding values (geBVs). Early estimates of the heritability of SM suggest it is around 0.7-0.8 (Preliminary estimate which may be subject to later modification) or that 70-80% of the variation between individuals is genetic in origin and about 20-30% is environmental. In the case of SM not much is known about the environmental influences and these may include in-utero or developmental effects. The heritability is sufficiently high, however, that genetic selection against the disease should be very successful. Heritabilities for Chiari Malformation, Cerebellar Herniation and Medullary Kinking are also very high. Genetic correlations between these traits and SM are positive and, interestingly, less than one. This suggests that different genes may be controlling SM and CM and that it will be possible to select against SM even if dogs have the malformation (CM).
One concern that we have at the present is that the estimates of heritability may be biased upwards. This is because the data has been ascertained on the basis of clinical cases. Most dogs will be MRI scanned because of concern that the dog may have SM or because the family or line is known to be afflicted by the condition. It is probably fairly rare that unaffected dogs from clear lines would be MRI scanned. We are taking various approaches to trying to iron out the bias, most based on modifying the statistical analysis, but it would also be beneficial if some dogs identified as unaffected could be MRI scanned. The plan is to then consolidate the estimated breeding values (EBVs) and try to estimate them for the entire UK registered population of CKCS. To help towards this aim we would like to collate the results of MRI scans coming from all clinics that are currently offering scans. We intend to set up a webpage where people can submit information directly and which gives details of where information can be sent by post. We hope to have this in operation in the next few weeks.
Estimated Breeding Values (EBVs) are the best measure available for complex traits of the genetic potential of individuals. The breeding value is the sum of the genetic effects and is the equivalent of an animal’s genotype at all the genes contributing to the disease. The EBVs for SM allow us to go from a dichotomous outcome (affected or unaffected) to an underlying continuous scale of liability. This gives us a much finer grading on which to evaluate dogs, leading to much more accurate selection. As an example, the figure below shows how EBVs compare with the A-F grading based on the MRI scans (gradings proposed by Clare Rusbridge). Grades A-C have more favourable (lower or more negative) EBVs while grades D-F are unfavourable. The gradings, however, do span a range of breeding values and some dogs graded A-C may have EBVs which suggest they can pass on a degree of disease risk to their offspring.
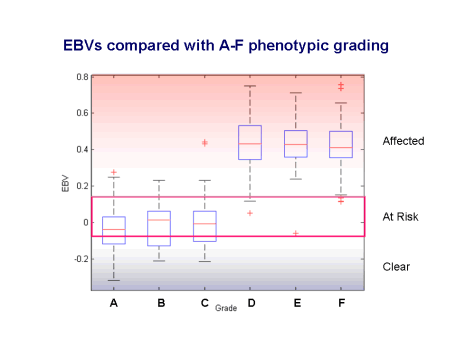 Using
EBVs allows us to distinguish between higher and lower risk dogs in the grading
categories A-C. EBVs can be calculated for most dogs even if they have not been
MRI scanned, as long as they are related to dogs that have been scanned. The
predicted EBV of an individual is half the EBV of its sire plus half the EBV of
its dam. All dogs will have an EBV at birth but the EBV may be modified by the
dog’s subsequent clinical record or MRI scan and by information coming from
other relatives.
Using
EBVs allows us to distinguish between higher and lower risk dogs in the grading
categories A-C. EBVs can be calculated for most dogs even if they have not been
MRI scanned, as long as they are related to dogs that have been scanned. The
predicted EBV of an individual is half the EBV of its sire plus half the EBV of
its dam. All dogs will have an EBV at birth but the EBV may be modified by the
dog’s subsequent clinical record or MRI scan and by information coming from
other relatives.
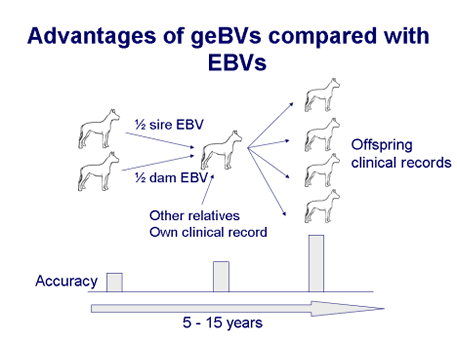
The EBV becomes more accurate as information on offspring becomes available, because we start to gain insight into which half of the sire and dam genes were actually inherited when we see transmission of the genes to offspring. The accuracy of the EBV increases with numbers of offspring and this may take some time to achieve. In contrast, genomic breeding values (geBVs) provide a high accuracy from birth. By looking directly at the DNA genotypes we can see which genes were inherited from the sire and from the dam, without having to wait for offspring. Genomic breeding values can be used for accurate evaluation at an early stage, before the disease phenotype may be apparent and before a dog is used for breeding.
 In
addition to selecting away from individual known diseases, such as
syringomyelia, it is important to consider the long-term health of the breed.
Population diversity and maintenance of diversity is important in order to
minimize the risk of future new diseases arising. We want to apply
state-of-the-art genetic selection techniques that use optimal contribution
theory to help avoid unequal representation of individuals in future generations
or ‘genetic bottlenecks’ occurring. This ensures that increases in inbreeding
and loss of diversity are minimized. Our aim is to develop internet-based tools
that allow breeders to have direct access to these state-of-the-art techniques
to help them make optimal selection decisions.
In
addition to selecting away from individual known diseases, such as
syringomyelia, it is important to consider the long-term health of the breed.
Population diversity and maintenance of diversity is important in order to
minimize the risk of future new diseases arising. We want to apply
state-of-the-art genetic selection techniques that use optimal contribution
theory to help avoid unequal representation of individuals in future generations
or ‘genetic bottlenecks’ occurring. This ensures that increases in inbreeding
and loss of diversity are minimized. Our aim is to develop internet-based tools
that allow breeders to have direct access to these state-of-the-art techniques
to help them make optimal selection decisions.
The Cavalier King Charles Spaniel will be the first dog breed in the world for which these techniques will be available. We also plan to explore different breeding strategies, based on computer modelling. This will help us to establish the time-scale over which the disease incidence can be reduced, the range of breeding values that should be used for breeding and the acceptable rate of diversity loss to minimize future disease risk. The immediate next steps in the project are to:
• Widen our data collection effort to include information for as broad a section of the population as we can. We will be setting up a webpage which will give details on how information can be submitted to us.
• Look at ways in which we can get a more accurate or unbiased estimate of the disease incidence.
• Investigate whether SM could be caused by a single gene or whether the multiple gene (polygenic) model fits the data better.
• Include information on Mitral Valve Disease (MVD) with the aim of producing EBVs for MVD.
We are also working towards the longer term aims of:
• Modelling different breeding strategies and identifying the most appropriate strategy.
• Carrying out molecular genetic analysis of SM and CM to identify the underlying genes, in collaboration with the University of Montreal, Clare Rusbridge and Penny Knowler.
• Developing genomic breeding values (geBVs) for SM.
Dr. Blott may be contacted at Genetics Department, Animal Health Trust, , Lanwades Park Kentford, Newmarket, Suffolk CB8 7UU, telephone +44 1638 751000, fax +44 1638 555606, website: www.aht.org.uk
RETURN TO TOP
Research News
 January 2015:
Research
shows limited genetic divergence between dog breeds in UK and South
Africa. In a
December 2014 report in the Veterinary Record, researchers (K. M.
Summers, R. Ogden, D. N. Clements, A. T. French, A. G. Gow, R. Powell,
B. Corcoran, R. J. Mellanby, and J. P. Schoeman) found very limited genetic
divergence between the dog breeds of the Labrador retriever, German
shepherd dog, and Jack Russell terrier in the geographically isolated
countries of the United Kingdom and South Africa. They attribute this to
similarity in breed standards, importation of dogs, and artificial
insemination. They concluded that:
January 2015:
Research
shows limited genetic divergence between dog breeds in UK and South
Africa. In a
December 2014 report in the Veterinary Record, researchers (K. M.
Summers, R. Ogden, D. N. Clements, A. T. French, A. G. Gow, R. Powell,
B. Corcoran, R. J. Mellanby, and J. P. Schoeman) found very limited genetic
divergence between the dog breeds of the Labrador retriever, German
shepherd dog, and Jack Russell terrier in the geographically isolated
countries of the United Kingdom and South Africa. They attribute this to
similarity in breed standards, importation of dogs, and artificial
insemination. They concluded that:
"An important implication of this study is that genetic diseases detected in a breed are likely to be found in both countries and could be transmitted from founders in either country, exacerbated by the use of artificial insemination, limiting the number of sires."
June
2013:
UK cavalier club and Kennel Club may revise CKCS breed standard for genetic
diversity.
 At the May 26 conference on genetic diversity in the
cavalier King Charles spaniel, sponsored by the UK Cavalier King Charles Spaniel
Club, Belgium's
Cavaliers for Life spokesman Arnold Jacques (right) recommended
that: "We could change the standard of the breed. If we omit the phrases [e.g.,
'White
markings undesirable'], that holds us back from making the combination between parti-colours and whole-colours, this could increase the genetic diversity of
the breed."
At the May 26 conference on genetic diversity in the
cavalier King Charles spaniel, sponsored by the UK Cavalier King Charles Spaniel
Club, Belgium's
Cavaliers for Life spokesman Arnold Jacques (right) recommended
that: "We could change the standard of the breed. If we omit the phrases [e.g.,
'White
markings undesirable'], that holds us back from making the combination between parti-colours and whole-colours, this could increase the genetic diversity of
the breed."
The UK publication, Dog World, reports that, following Mr. Jacques' presentation, the cavalier club proposed to the CKCS regional clubs that changes be made to the breed standard so that references to "white undesirable" are removed from the whole colors, and the words "broken up" in the phrase "black and white well spaced, broken up…" removed from the particolors. The UK Kennel Club's health and breeder services manager, Bill Lambert, who was present at the conference, said that if the majority of cavalier clubs gave their support to the revision of the breed standard, the Kennel Club would be "highly likely" to make the changes. "How much more attractive than outcrossing!" he said.
May 2013: UK Kennel Club announces syringomyelia as a "candidate" for EBVs. The UK Kennel Club, in its 2012 Dog Health Group Annual Report, announced that syringomyelia (along with mitral valve disease) "are candidates for the development of EBVs but require appropriate data collection procedures to be in place." The report goes on:
"A BVA/KC scheme for syringomyelia was launched in 2012 and once enough data has accumulated through the scheme then EBVs for the condition will become a possibility."
 July
2012: French researchers
confirm the obvious in trying to control inherited disorders and genetic
diversity in dogs. French geneticists Grégoire Leroy
and Xavier Rognon
report in the Veterinary
Journal that:
July
2012: French researchers
confirm the obvious in trying to control inherited disorders and genetic
diversity in dogs. French geneticists Grégoire Leroy
and Xavier Rognon
report in the Veterinary
Journal that:
"Breeding policies, such as the removal of all carriers from the reproduction pool, may have a range of effects on genetic diversity, depending on the breed and the frequency of deleterious alleles. Limiting the number of offspring per reproducer may also have a positive impact on genetic diversity."
May 2012: Belgium & Dutch cavalier group announces EBV pedigree database. "Cavaliers for Life", an ambitious group of Belgium and Dutch cavalier fanciers, led by Arnold Jacques and Pauline Jordens, has announced that their group's database now contains the pedigrees of all the cavaliers born in the last fifteen years in Holland and Belgium. They stated that, from that data, The University of Leuven in Belgium has carried out a full study of the genetic variation and inbreeding within the breed in Belgium and Holland.
RETURN TO TOP
Related Links
RETURN TO TOP
Veterinary Resources
Coefficients of Inbreeding and Relationship. Sewall Wright. American Naturalist. July 1922;56(645):330-338.
The
things we do to dogs. Simon Wolfensohn. New Scientist. May
1981;90.1253:404-407. Quote: "Man is apt to refer to dog as his best
friend, but it's doubtful whether dog can return the compliment. Our
strange ideas about the appearance of our canine companions have
afflicted many breeds with physical deformities which at best cause the
dogs considerable inconvenience and at worst result in a lifetime of
visits to the vet. Some breeds have developed their own
![]() hereditary
diseases because their short-sighted breeden have been more concerned
with their appearance than their general state of health. Not content
with breeding dogs into peculiar shapes and sizes, we have even taken to
snipping off bits of their anatomy to make them conform to our misguided
ideals." (Watch Dr. Simon Wolfensohn discuss "Pedigree dog health
problems" at Crufts in 1985
on
YouTube.)
hereditary
diseases because their short-sighted breeden have been more concerned
with their appearance than their general state of health. Not content
with breeding dogs into peculiar shapes and sizes, we have even taken to
snipping off bits of their anatomy to make them conform to our misguided
ideals." (Watch Dr. Simon Wolfensohn discuss "Pedigree dog health
problems" at Crufts in 1985
on
YouTube.)
Genetic variability in French dog breeds assessed by pedigree data. G. Leroy, X. Rognon, A. Varlet, C. Joffrin, & E. Verrier. J. Anim. Breed. Genet. (Jan. 2006) 123:1–9. "To conclude, probabilities of gene origin added to computation of inbreeding gave us a lot of information about genetic structure and differences among breeds. Such results can be used to manage the genetic variability of breeds. For the breeds with a large population size, it is possible to limit the number of litters per male: such a rule is already applied in the German Shepherd dog in Germany (Guyader 1989). Moreover breed clubs could support breeding animals, which are little related to the whole population. Such dogs can be detected for instance by the average relatedness of each individual with the whole population (Goyache et al. 2003). For breeds with a small and/or decreasing population size, mating should be made between little related dogs."
Population Structure and Inbreeding From Pedigree Analysis of Purebred Dogs. Federico C. F. Calboli, Jeff Sampson, Neale Fretwell, and David J. Balding. Genetics May 2008;179(1):593-601. Quote: "Dogs are of increasing interest as models for human diseases, and many canine population-association studies are beginning to emerge. The choice of breeds for such studies should be informed by a knowledge of factors such as inbreeding, genetic diversity, and population structure, which are likely to depend on breed-specific selective breeding patterns. To address the lack of such studies we have exploited one of the world's most extensive resources for canine population-genetics studies: the United Kingdom (UK) Kennel Club registration database. We chose 10 representative breeds and analyzed their pedigrees since electronic records were established around 1970, corresponding to about eight generations before present. We find extremely inbred dogs in each breed except the greyhound and estimate an inbreeding effective population size between 40 and 80 for all but 2 breeds. For all but 3 breeds, >90% of unique genetic variants are lost over six generations, indicating a dramatic effect of breeding patterns on genetic diversity. We introduce a novel index Ψ for measuring population structure directly from the pedigree and use it to identify subpopulations in several breeds. As well as informing the design of canine population genetics studies, our results have implications for breeding practices to enhance canine welfare. ... We have found that the loss of genetic diversity is very high, with many breeds losing >90% of singleton variants in just six generations. ... Dog breeds are required to conform to a breed standard, the pursuit of which often involves intensive inbreeding ... This has adverse consequences in terms of loss of genetic variability and high prevalence of recessive genetic disorders. These features make purebred dogs attractive for the study of genetic disorders, but raise concerns about canine welfare. ... On the basis of these results, we concur with Leroy et al. (2006) that remedial action to maintain or increase genetic diversity should now be a high priority in the interests of the health of purebred dogs. Possible remedial action includes limits on the use of popular sires, encouragement of matings across national and continental boundaries, and even the relaxation of breed rules to permit controlled outcrossing."
Inherited defects in pedigree dogs. Part 1: Disorders related to breed standards. Lucy Asher, Gillian Diesel, Jennifer F. Summers, Paul D. McGreevy, Lisa M. Collins. Vet. J. December 2009;182(3):402-411. Quote: "The United Kingdom pedigree-dog industry has faced criticism because certain aspects of dog conformation stipulated in the UK Kennel Club breed standards have a detrimental impact on dog welfare. A review of conformation-related disorders was carried out in the top 50 UK Kennel Club registered breeds using systematic searches of existing information. A novel index to score severity of disorders along a single scale was also developed and used to conduct statistical analyses to determine the factors affecting reported breed predisposition to defects. [Cavalier King Charles spaniel: total disorders: 25.] ... Some large breeds are also reportedly predisposed to valvular disease (e.g. German Shepherd Dog), however, mitral valve disease (severity: 7–12) is more commonly associated with small dog breeds (Borgarelli et al., 2004), particularly the Cavalier King Charles spaniel. The prevalence estimates for mitral valve disease in the Cavalier King Charles spaniel range between 11–45% (Hyun, 2005; Haggstrom et al., 1992; Darke, 1987). ... According to the literature searched, each of the top 50 breeds was found to have at least one aspect of its conformation predisposing it to a disorder; and 84 disorders were either directly or indirectly associated with conformation. The Miniature poodle, Bulldog, Pug and Basset hound had most associations with conformation-related disorders. Further research on prevalence and severity is required to assess the impact of different disorders on the welfare of affected breeds."
Heritability of syringomyelia in Cavalier King Charles spaniels. Tom Lewis, Clare Rusbridge, Penny Knowler, Sarah Blott, John A. Woolliams. Vet.J. March 2010;183(3): 345-347. Quote: "Mixed model analysis of 384 Cavalier King Charles spaniels (CKCS), with a magnetic resonance imaging diagnosis for the presence or absence of a syrinx, in conjunction with the Kennel Club pedigree records of all dogs registered from the mid 1980s to September 2007, revealed a moderately high estimate of heritability of syringomyelia (h2 = 0.37 ± 0.15 standard error) when analysed as a binary trait. Inspection of cases where the disease segregated within families pointed to genes at more than one locus influencing syringomyelia. The availability of estimated breeding values for Kennel Club registered CKCS is a significant step in being able to select against syringomyelia, particularly given the difficulty of ascertaining the disease phenotype."
Localization of Canine Brachycephaly Using an Across Breed Mapping Approach. Danika Bannasch, Amy Young, Jeffrey Myers, Katarina Truvé, Peter Dickinson, Jeffrey Gregg, Ryan Davis, Eric Bongcam-Rudloff, Matthew T. Webster, Kerstin Lindblad-Toh, Niels Pedersen. PLOS One. March 2010. Quote: "The domestic dog, Canis familiaris, exhibits profound phenotypic diversity and is an ideal model organism for the genetic dissection of simple and complex traits. However, some of the most interesting phenotypes are fixed in particular breeds and are therefore less tractable to genetic analysis using classical segregation-based mapping approaches. We implemented an across breed mapping approach using a moderately dense SNP array, a low number of animals and breeds carefully selected for the phenotypes of interest to identify genetic variants responsible for breed-defining characteristics. Using a modest number of affected (10–30) and control (20–60) samples from multiple breeds, the correct chromosomal assignment was identified in a proof of concept experiment using three previously defined loci; hyperuricosuria, white spotting and chondrodysplasia. Genome-wide association was performed in a similar manner for one of the most striking morphological traits in dogs: brachycephalic head type. Although candidate gene approaches based on comparable phenotypes in mice and humans have been utilized for this trait, the causative gene has remained elusive using this method. Samples from nine affected breeds and thirteen control breeds identified strong genome-wide associations for brachycephalic head type on Cfa 1. Two independent datasets identified the same genomic region. Levels of relative heterozygosity in the associated region indicate that it has been subjected to a selective sweep, consistent with it being a breed defining morphological characteristic. Genotyping additional dogs in the region confirmed the association. To date, the genetic structure of dog breeds has primarily been exploited for genome wide association for segregating traits. These results demonstrate that non-segregating traits under strong selection are equally tractable to genetic analysis using small sample numbers."
Optimisation of breeding strategies to reduce the prevalence of inherited disease in pedigree dogs. Lewis, T.W.; Woolliams, J.A.; Blott, S.C. Animal Welfare 19(Supp 1):93-98(6), May 2010. Quote: "One option for improving the welfare of purebred dog breeds is to implement health breeding programmes, which allow selection to be directed against known diseases while controlling the rate of inbreeding to a minimal level in order to maintain the long-term health of the breed. The aim of this study is to evaluate the predicted impact of selection against disease in two breeds: the Cavalier King Charles spaniel (CKCS) .... Heritabilities for mitral valve disease, syringomyelia in the CKCS ... were estimated to be 0.64 (± 0.07), 0.32 (± 0.125) ... respectively, which suggest encouraging selection responses are feasible based upon the estimation of breeding values (EBVs) if monitoring schemes are maintained for these breeds. Although using data from disease databases can introduce problems due to bias, as a result of individuals and families with disease usually being over-represented, the data presented is a step forward in providing information on risk. EBVs will allow breeders to distinguish between potential parents of high and low risk, after removing the influence of life history events. Analysis of current population structure, including numbers of dogs used for breeding, average kinship and average inbreeding provides a basis from which to compare breeding strategies. Predictions can then be made about the number of generations it will take to eradicate disease, the number of affected individuals that will be born during the course of selective breeding and the benefits that can be obtained by using optimisation to constrain inbreeding to a pre-defined sustainable rate.
Mating practices and the dissemination of genetic disorders in domestic animals, based on the example of dog breeding. Leroy G, Baumung R. Anim Genet. February 2011;42(1):66-74. Quote: On the basis of simulations and genealogical data of ten dog breeds, three popular mating practices (popular sire effect, line breeding, close breeding) were investigated along with their effects on the dissemination of genetic disorders. Our results showed that the use of sires in these ten breeds is clearly unbalanced. Depending on the breed, the effective number of sires represented between 33% and 70% of the total number of sires. Mating between close relatives was also found to be quite common, and the percentage of dogs inbred after two generations ranged from 1% to about 8%. A more or less long-term genetic differentiation, linked to line breeding practices, was also emphasized in most breeds. F(IT) index based on gene dropping proved to be efficient in differentiating the effects of the different mating practices, and it ranged from -1.3% to 3.2% when real founders were used to begin a gene dropping process. Simulation results confirmed that the popular sire practice leads to a dissemination of genetic disorders. Under a realistic scenario, regarding the imbalance in the use of sires, the dissemination risk was indeed 4.4 times higher than under random mating conditions. In contrast, line breeding and close breeding practices tend to decrease the risk of the dissemination of genetic disorders.
Getting priorities straight: risk assessment and decision-making in the improvement of inherited disorders in pedigree dogs. Collins LM, Asher L, Summers J, McGreevy P. Vet J. 2011 Aug;189(2):147-54. Quote: "The issue of inherited disorders in pedigree dogs is not a recent phenomenon and reports of suspected genetic defects associated with breeding practices date back to Charles Darwin's time. In recent years, much information on the array of inherited defects has been assimilated and the true extent of the problem has come to light. Historically, the direction of research funding in the field of canine genetic disease has been largely influenced by the potential transferability of findings to human medicine, economic benefit and importance of dogs for working purposes. More recently, the argument for a more canine welfare-orientated approach has been made, targeting research efforts at the alleviation of the most suffering in the greatest number of animals. A method of welfare risk assessment was initially developed as a means of objectively comparing, and thus setting priorities for, different welfare problems. The method has been applied to inherited disorders in pedigree dogs to investigate which disorders have the greatest welfare impact and which breeds are most affected. Work in this field has identified 396 inherited disorders in the top 50 most popular breeds in the UK. This article discusses how the results of welfare risk assessment for inherited disorders can be used to develop strategies for improving the health and welfare of dogs in the long term. A new risk assessment criterion, the Breed-Disorder Welfare Impact Score (BDWIS), which takes into account the proportion of life affected by a disorder, is introduced. A set of health and welfare goals is proposed and strategies for achieving these goals are highlighted, along with potential rate-determining factors at each step."
Genetic diversity, inbreeding and breeding practices in dogs: results from pedigree analyses. Leroy G. Vet J. 2011 Aug;189(2):177-82. Quote: "Pedigree analysis constitutes a classical approach for the study of the evolution of genetic diversity, genetic structure, history and breeding practices within a given breed. As a consequence of selection pressure, management in closed populations and historical bottlenecks, many dog breeds have experienced considerable inbreeding and show (on the basis of a pedigree approach) comparable diversity loss compared to other domestic species. This evolution is linked to breeding practices such as the overuse of popular sires or mating between related animals. The popular sire phenomenon is the most problematic breeding practice, since it has also led to the dissemination of a large number of inherited defects. The practice should be limited by taking measures such as restricting the number of litters (or offspring) per breeding animal."
Inbreeding and genetic diversity in dogs: results from DNA analysis. Wade CM. Vet J. 2011 Aug;189(2):183-8. Quote: "This review assesses evidence from DNA analysis to determine whether there is sufficient genetic diversity within breeds to ensure that populations are sustainable in the absence of cross breeding and to determine whether genetic diversity is declining. On average, dog breeds currently retain approximately 87% of the available domestic canine genetic diversity. Requirements that breeding stock must be 'clear' for all genetic disorders may firstly place undue genetic pressure on animals tested as being 'clear' of known genetic disorders, secondly may contribute to loss of diversity and thirdly may result in the dissemination of new recessive disorders for which no genetic tests are available. Global exchange of genetic material may hasten the loss of alleles and this practice should be discussed in relation to the current effective population size of a breed and its expected future popularity. Genomic data do not always support the results from pedigree analysis and possible reasons for this are discussed."
Is gene loss in pedigree dogs surprisingly rapid? James JW. Vet J. August 2011;189(2):211-3. Quote: Factors affecting the probabilities of gene loss are discussed, with particular attention given to population expansion, sex ratio and inbreeding. Much of the variation in gene survival probabilities among breeds can be explained by differences in expansion rate, sex ratio and family size, with little or no influence of average inbreeding and population size.
Researcher responsibilities and genetic counseling for pure-bred dog populations. Bell Jerold S. Vet J. 2011 Aug;189(2):234-5. Quote: "Breeders of dogs have ethical responsibilities regarding the testing and management of genetic disease. Molecular genetics researchers have their own responsibilities, highlighted in this article. Laboratories offering commercial genetic testing should have proper sample identification and quality control, official test result certificates, clear explanations of test results and reasonably priced testing fees. Providing test results to a publicly-accessible genetic health registry allows breeders and the public to search for health-tested parents to reduce the risk of producing or purchasing affected offspring. Counseling on the testing and elimination of defective genes must consider the effects of genetic selection on the population. Recommendations to breed quality carriers to normal-testing dogs and replacing them with quality normal-testing offspring will help to preserve breeding lines and breed genetic diversity."
Genetic Evaluation of Hip Score in UK Labrador Retrievers. Thomas W. Lewis, Sarah C. Blott, John A. Woolliams. PLoS ONE 5(10): e12797. . Quote: "The presented results have demonstrated that the availability of EBV through routine evaluations of the hip score data would hasten progress in alleviating the problem of hip dysplasia via increases in selective accuracy compared to selection based on phenotype alone. However the benefits of EBV extend beyond the simple comparisons of accuracy for a recently scored dog: (i) the EBV for an individual, unlike its phenotypic score, will further increase in accuracy over time by utilising all the available information and being updated as additional information becomes available e.g. from offspring or siblings; (ii) the EBV will provide predictors for those animals that do not have a phenotypic record hence increasing selection opportunities and intensity, which again enhances rate of improvement; (iii) the EBV will be available from the moment of birth for selection (although newborn littermates will have identical EBV) and, in this case, the accuracy (and hence rate of improvement) from using EBV increases by 31% compared to the parental average phenotype; (iv) the EBV will have been corrected for other fixed effects such as sex and age which bias phenotype as a predictor of genetic merit; and (v) it may be argued that taking account of a sustainable rate of inbreeding as well as disease prevalence would restrict the selection pressure that can be applied, however this only serves to place a greater emphasis on the accuracy of the selection that does take place. Finally with the availability of sequence [24] and dense canine SNP chips, the development of a genomic EBV (an EBV informed by additional information from dense SNP genotypes [25]) would help to distinguish littermates and further increase accuracy, increasing the potential rate of improvement, and might also lead subsequently to scientific benefits through identifying the major QTL. The intention is to make public the EBV for hip score for all KC registered Labrador Retrievers so that all these benefits may be realised. "
Identification of Genomic Regions Associated with Phenotypic Variation between Dog Breeds using Selection Mapping. Amaury Vaysse, Abhirami Ratnakumar, Thomas Derrien, Erik Axelsson, Gerli Rosengren Pielberg, Snaevar Sigurdsson, Tove Fall, Eija H. Seppälä, Mark S. T. Hansen, Cindy T. Lawley, Elinor K. Karlsson, The LUPA Consortium, Danika Bannasch, Carles Vilà, Hannes Lohi, Francis Galibert, Merete Fredholm, Jens Häggström, Åke Hedhammar, Catherine André, Kerstin Lindblad-Toh, Christophe Hitte, Matthew T. Webster. October 2011. PLOS Genetics. Quote: The extraordinary phenotypic diversity of dog breeds has been sculpted by a unique population history accompanied by selection for novel and desirable traits. Here we perform a comprehensive analysis using multiple test statistics to identify regions under selection in 509 dogs from 46 diverse breeds using a newly developed high-density genotyping array consisting of >170,000 evenly spaced SNPs. We first identify 44 genomic regions exhibiting extreme differentiation across multiple breeds. Genetic variation in these regions correlates with variation in several phenotypic traits that vary between breeds, and we identify novel associations with both morphological and behavioral traits. We next scan the genome for signatures of selective sweeps in single breeds, characterized by long regions of reduced heterozygosity and fixation of extended haplotypes. These scans identify hundreds of regions, including 22 blocks of homozygosity longer than one megabase in certain breeds. Candidate selection loci are strongly enriched for developmental genes. We chose one highly differentiated region, associated with body size and ear morphology, and characterized it using high-throughput sequencing to provide a list of variants that may directly affect these traits. This study provides a catalogue of genomic regions showing extreme reduction in genetic variation or population differentiation in dogs, including many linked to phenotypic variation. The many blocks of reduced haplotype diversity observed across the genome in dog breeds are the result of both selection and genetic drift, but extended blocks of homozygosity on a megabase scale appear to be best explained by selection. Further elucidation of the variants under selection will help to uncover the genetic basis of complex traits and disease.
Empowering international canine inherited disorder management. Bethany J. Wilson and Claire M. Wade. Mammalian Genome. October 2011. Quote: For multifactorial traits, high-quality genetic evaluations are typically conducted by scoring one or more appropriate phenotypes and calculating Estimated Breeding Values (EBVs) for the disorder ... An evaluated animal’s EBV represents the superiority of the animal’s genes that affect the breeding objective relative to the complement of genes of the ‘‘average’’ animal in the breed or in a cohort. All available information about the animal’s value as a breeding animal is combined, with appropriate statistical weighting techniques, into the EBV. When considering disease phenotypes, the information from the candidate itself, its relatives, marker genotypes related to the phenotype from either the individual of concern or its relatives, and phenotypes or marker genotypes for genetically correlated traits (traits controlled by common genetic loci) can be incorporated into the evaluation process. The quality of the information provided by each information source in consideration of the breeding objective affects the importance accorded this information in the final EBV calculation. ... Because in dog populations relatively few progeny are derived from any given parent, it may not be possible to evaluate entirely genomic EBVs for dogs with great accuracy ... and so the collection, storage, cleaning, and processing of phenotype and pedigree information will remain necessary for the foreseeable future. ... There is considerable current research activity toward understanding the genes responsible for single-locus genetic disorders and the genes that contribute in a large way to complex genetic disorders. This research is vital for understanding the aetiology and pathogenesis of these disorders, providing a foundation for future development of treatments and therapies, understanding the diseases as a potential model for analogous human disease and the development of genetic tests for these genes (or markers of these genes) to potentially aid control through selective breeding. Generally, it is the last of these aims, genetic tests, which has the greatest potential for immediate improvement of canine welfare.
Assessing the impact of breeding strategies on inherited disorders and genetic diversity in dogs. Grégoire Leroy, Xavier Rognon. Vet.J. July 2012. Quote: "In the context of management of genetic diversity and control of genetic disorders within dog breeds, a method is proposed for assessing the impact of different breeding strategies that takes into account the genealogical information specific to a given breed. Two types of strategies were investigated: (1) eradication of an identified monogenic recessive disorder, taking into account three different mating limitations and various initial allele frequencies; and (2) control of the population sire effect by limiting the number of offspring per reproducer. The method was tested on four dog breeds: Braque Saint Germain, Berger des Pyrénées, Coton de Tulear and Epagneul Breton. Breeding policies, such as the removal of all carriers from the reproduction pool, may have a range of effects on genetic diversity, depending on the breed and the frequency of deleterious alleles. Limiting the number of offspring per reproducer may also have a positive impact on genetic diversity."
So many doggone traits: mapping genetics of multiple phenotypes in the domestic dog. Maud Rimbault and Elaine A. Ostrander. Human Molecular Genetics. August 2012. Quote: "The worldwide dog population is fragmented into >350 domestic breeds. Breeds share a common ancestor, the gray wolf. The intense artificial selection imposed by humans to develop breeds with particular behaviors and phenotypic traits has occurred primarily in the last 200–300 years. As a result, the number of genes controlling the major differences in body size, leg length, head shape, etc. that define each dog is small, and genetically tractable. This is in comparison to many human complex traits where small amounts of variance are controlled by literally hundreds of genes. We have been interested in disentangling the genetic mechanisms controlling breed-defining morphological traits in the domestic dog. The structure of the dog population, comprised large numbers of pure breeding populations, makes this task surprisingly doable. In this review, we summarize recent work on the genetics of body size, leg length and skull shape, while setting the stage for tackling other traits. It is our expectation that these results will contribute to a better understanding of mammalian developmental processes overall."
Variation of BMP3 Contributes to Dog Breed Skull Diversity. Jeffrey J. Schoenebeck, Sarah A. Hutchinson, Alexandra Byers, Holly C. Beale, Blake Carrington, Daniel L. Faden, Maud Rimbault, Brennan Decker, Jeffrey M. Kidd, Raman Sood, Adam R. Boyko, John W. Fondon III, Robert K. Wayne, Carlos D. Bustamante, Brian Ciruna, Elaine A. Ostrander. PLOS Genetics. August 2012. Quote: "Since the beginnings of domestication, the craniofacial architecture of the domestic dog has morphed and radiated to human whims. By beginning to define the genetic underpinnings of breed skull shapes, we can elucidate mechanisms of morphological diversification while presenting a framework for understanding human cephalic disorders. Using intrabreed association mapping with museum specimen measurements, we show that skull shape is regulated by at least five quantitative trait loci (QTLs). Our detailed analysis using whole-genome sequencing uncovers a missense mutation in BMP3. Validation studies in zebrafish show that Bmp3 function in cranial development is ancient. Our study reveals the causal variant for a canine QTL contributing to a major morphologic trait."
Do dog owners perceive the clinical signs related to conformational inherited disorders as ‘normal’ for the breed? A potential constraint to improving canine welfare. (Owner recognition of breathing disorders in brachycephalic dogs). RMA Packer, A Hendricks and CC Burn. Animal Welfare. 2012;21(S1):81-93. Quote: "Selection for brachycephalic (foreshortened muzzle) phenotypes in dogs is a major risk factor for brachycephalic obstructive airway syndrome (BOAS). Clinical signs include respiratory distress, exercise intolerance, upper respiratory noise and collapse. Efforts to combat BOAS may be constrained by a perception that it is ‘normal’ in brachycephalic dogs. This study aimed to quantify ownerperception of the clinical signs of BOAS as a veterinary problem. A questionnaire-based study was carried out over five months on the owners of dogs referred to the Queen Mother Hospital for Animals (QMHA) for all clinical services, except for Emergency and Critical Care. Owners reported the frequency of respiratory difficulty and characteristics of respiratory noise in their dogs in four scenarios, summarised as an ‘owner-reported breathing’ (ORB) score. Owners then reported whether their dog currently has, or has a history of, ‘breathing problems’. Dogs (n = 285) representing 68 breeds were included, 31 of which were classed as ‘affected’ by BOAS either following diagnostics, or by fitting case criteria based on their ORB score, skull morphology and presence of stenotic nares. The median ORB score given by affected dogs’ owners was 20/40 (range 8–30). Over half (58%) of owners of affected dogs reported that their dog did not have a breathing problem. This marked disparity between owners’ reports of frequent, severe clinical signs and their perceived lack of a ‘breathing problem’ in their dogs is of concern. Without appreciation of the welfare implications of BOAS, affected but undiagnosed dogs may be negatively affected indefinitely through lack of treatment. Furthermore, affected dogs may continue to be selected in breeding programmes, perpetuating this disorder."
Breed Health Improvement Strategy: a Step-by-step Guide. Ian Seath. The Kennel Club. October 2012.
The effects of dog breed development on genetic diversity and the relative influences of performance and conformation breeding. N. Pedersen, H. Liu, G. Theilen, B. Sacks. Anim.Breeding&Genetics. December 2012. Quote: "Genetic diversity was compared among eight dog breeds selected primarily for conformation (Standard Poodle, Italian Greyhound and show English Setter), conformation and performance (Brittany), predominantly performance (German Shorthaired and Wirehaired Pointers) or solely performance (field English Setter and Red Setter). Modern village dogs, which better reflect ancestral genetic diversity, were used as the standard. Four to seven maternal and one to two Y haplotypes were found per breed, with one usually dominant. Diversity of maternal haplotypes was greatest in village dogs, intermediate in performance breeds and lowest in conformation breeds. Maternal haplotype sharing occurred across all breeds, while Y haplotypes were more breed specific. Almost all paternal haplotypes were identified among village dogs, with the exception of the dominant Y haplotype in Brittanys, which has not been identified heretofore. The highest heterozygosity based on 24 autosomal microsatellites was found in village dogs and the lowest in conformation (show) breeds. Principal coordinate analysis indicated that conformation-type breeds were distinct from breeds heavily used for performance, the latter clustering more closely with village dogs. The Brittany, a well-established dual show and field breed, was also genetically intermediate between the conformation and performance breeds. The number of DLA-DRB1 alleles varied from 3 to 10 per breed with extensive sharing. SNPs across the wider DLA region were more frequently homozygous in all pure breeds than in village dogs. Compared with their village dog relatives, all modern breed dogs exhibit reduced genetic diversity. Genetic diversity was even more reduced among breeds under selection for show/conformation."
Comparative analyses of genetic trends and prospects for selection against hip and elbow dysplasia in 15 UK dog breeds. Lewis TW, Blott SC, Woolliams JA. BMC Genet. March 2013;14:16. Quote: "Background: Hip dysplasia remains one of the most serious hereditary diseases occurring in dogs despite long-standing evaluation schemes designed to aid selection for healthy joints. Many researchers have recommended the use of estimated breeding values (EBV) to improve the rate of genetic progress from selection against hip and elbow dysplasia (another common developmental orthopaedic disorder), but few have empirically quantified the benefits of their use. This study aimed to both determine recent genetic trends in hip and elbow dysplasia, and evaluate the potential improvements in response to selection that publication of EBV for such diseases would provide, across a wide range of pure-bred dog breeds. Results: The genetic trend with respect to hip and elbow condition due to phenotypic selection had improved in all breeds, except the Siberian Husky. However, derived selection intensities are extremely weak, equivalent to excluding less than a maximum of 18% of the highest risk animals from breeding. EBV for hip and elbow score were predicted to be on average between 1.16 and 1.34 times more accurate than selection on individual or both parental phenotypes. Additionally, compared to the proportion of juvenile animals with both parental phenotypes, the proportion with EBV of a greater accuracy than selection on such phenotypes increased by up to 3-fold for hip score and up to 13-fold for elbow score. Conclusions: EBV are shown to be both more accurate and abundant than phenotype, providing more reliable information on the genetic risk of disease for a greater proportion of the population. Because the accuracy of selection is directly related to genetic progress, use of EBV can be expected to benefit selection for the improvement of canine health and welfare. Public availability of EBV for hip score for the fifteen breeds included in this study will provide information on the genetic risk of disease in nearly a third of all dogs annually registered by the UK Kennel Club, with in excess of a quarter having an EBV for elbow score as well."
Estimated Breeding Values for Canine Hip Dysplasia Radiographic Traits in a Cohort of Australian German Shepherd Dogs. Bethany J. Wilson, Frank W. Nicholas, John W. James, Claire M. Wade, and Peter C. Thomson. PLoS One. October 2013;8(10):e77470. Quote: "Canine hip dysplasia (CHD) is a serious and common musculoskeletal disease of pedigree dogs and therefore represents both an important welfare concern and an imperative breeding priority. The typical heritability estimates for radiographic CHD traits suggest that the accuracy of breeding dog selection could be substantially improved by the use of estimated breeding values (EBVs) in place of selection based on phenotypes of individuals. The British Veterinary Association/Kennel Club scoring method is a complex measure composed of nine bilateral ordinal traits, intended to evaluate both early and late dysplastic changes. However, the ordinal nature of the traits may represent a technical challenge for calculation of EBVs using linear methods. The purpose of the current study was to calculate EBVs of British Veterinary Association/Kennel Club traits in the Australian population of German Shepherd Dogs, using linear (both as individual traits and a summed phenotype), binary and ordinal methods to determine the optimal method for EBV calculation. Ordinal EBVs correlated well with linear EBVs (r=0.90–0.99) and somewhat well with EBVs for the sum of the individual traits (r=0.58–0.92). Correlation of ordinal and binary EBVs varied widely (r=0.24–0.99) depending on the trait and cut-point considered. The ordinal EBVs have increased accuracy (0.48–0.69) of selection compared with accuracies from individual phenotype-based selection (0.40–0.52). Despite the high correlations between linear and ordinal EBVs, the underlying relationship between EBVs calculated by the two methods was not always linear, leading us to suggest that ordinal models should be used wherever possible. As the population of German Shepherd Dogs which was studied was purportedly under selection for the traits studied, we examined the EBVs for evidence of a genetic trend in these traits and found substantial genetic improvement over time. This study suggests the use of ordinal EBVs could increase the rate of genetic improvement in this population."
Limited genetic divergence between dog breeds from geographically isolated countries. K. M. Summers, R. Ogden, D. N. Clements, A. T. French, A. G. Gow, R. Powell, B. Corcoran, R. J. Mellanby, J. P. Schoeman. Vet. Record. December 2014;175(22). Quote: "Genetic isolation in a population of UK dogs (Mellanby and others 2013) has been demonstrated previously. ... The authors were interested to assess whether dog breeds in another country had similar levels of genetic isolation. ... Genotypes using 15 microsatellite markers were established for 14 labrador retrievers, 26 German shepherd dogs and 35 Jack Russell terriers from South Africa, using the previously published protocol (Ogden and others 2011, Mellanby and others 2013). Calculations of heterozygosity and relative genetic distance were performed ... This analysis showed that within a breed the dogs were genetically very similar regardless of the country. Jack Russell terriers showed equal contributions from different subpopulations, whereas labrador retrievers and German shepherd dogs each showed 90 per cent contribution from a single subpopulation. Jack Russell terriers had very little contribution from either of the subpopulations predominant in the labrador retrievers or German shepherd dogs, as might be expected given the very different phenotype and ancestry of these dogs. The admixture of subpopulations within a breed was the same regardless of country, showing that the breeds have not diverged greatly. ... This analysis shows that the assignment of dogs to pedigree classes on the basis of genotype is robust across the two countries and there is little evidence for genetic drift within breeds, reflecting the similarity of the breed standards between the two countries. This may be maintained by the import of dogs from other countries and the exchange of semen for artificial insemination. ... Thus, the geographical isolation of the two countries has not apparently led to genetic divergence within the breeds examined, suggesting that there is minimal reproductive isolation and considerable genetic exchange. Consequently, an important implication of this study is that genetic diseases detected in a breed (Asher and others 2009, Summers and others 2010) are likely to be found in both countries and could be transmitted from founders in either country, exacerbated by the use of artificial insemination, limiting the number of sires."
The challenges of pedigree dog health: approaches to combating inherited disease. Lindsay L Farrell, Jeffrey J Schoenebeck, Pamela Wiener, Dylan N Clements, Kim M Summers. Canine Genetics & Epidemiology. February 2015;2(3):1-14. Quote: "The issue of inherited disorders and poor health in pedigree dogs has been widely discussed in recent years. With the advent of genome-wide sequencing technologies and the increasing development of new diagnostic DNA disease tests, the full extent and prevalence of inherited disorders in pedigree dogs is now being realized. In this review we discuss the challenges facing pedigree dog breeds: the common pitfalls and problems associated with combating single gene mediated disorders, phenotypic selection on complex disorders, and ways of managing genetic diversity. ... The Cavalier King Charles Spaniel (CKCS) breed is susceptible to 25 inherited disorders, the most common of which is early-onset myxomatous mitral valve disease (MMVD). In a 2004 survey, 42.8% of all UK CKCS died due to cardiac causes and there is increasing evidence that CKCS mitral valve disease is genetic in origin, with a heritability of between 0.33 and 0.67. ... Breeding strategies incorporating screening schemes have been shown to be successful in significantly reducing the prevalence of an inherited disorder and improving the overall health in certain breeds. However, with 215 breeds officially recognized by the Kennel Club in the United Kingdom and 396 inherited disorders currently identified, many breeds have reached the point at which successfully breeding away from susceptible individuals at a population-wide scale will require new genomic selection strategies in combination with currently available breeding schemes. Whilst DNA-based tests identifying disease causing mutation(s) remain the most informative and effective approach for single gene disorder disease management, they must be used along with current screening schemes, genomic selection, and pedigree information in breeding programs in the effort to maintain genetic diversity while also significantly reducing the number of inherited disorders in pedigree dogs."
Trends in genetic diversity for all Kennel Club registered pedigree dog breeds. T. W. Lewis, B.M. Abhayaratne, S. C. Blott. Canine Genetics and Epidemiology. September 2015;2:13. Quote: "Background: Inbreeding is inevitable in closed populations with a finite number of ancestors and where there is selection. Therefore, management of the rate of inbreeding at sustainable levels is required to avoid the associated detrimental effects of inbreeding. Studies have shown some pedigree dog breeds to have high levels of inbreeding and a high burden of inherited disease unrelated to selection objectives, implying loss of genetic diversity may be a particular problem for pedigree dogs. Pedigree analysis of all 215 breeds currently recognised by the UK Kennel Club over the period 1980–2014 was undertaken to ascertain parameters describing the rate of loss of genetic diversity due to inbreeding, and the presence of any general trend across all breeds. Results: The trend over all breeds was for the rate of inbreeding to be highest in the 1980s and 1990s, tending to decline after 2000. The trend was comparable in very common and rarer breeds, although was more pronounced in rarer breeds. Rates of inbreeding over the entire period 1980–2014 were not correlated with census population size. The existence of popular sires was apparent in all breeds. Conclusion: The trends detected over 1980–2014 imply an initial excessive loss of genetic diversity which has latterly fallen to sustainable levels, even with modest restoration in some cases. The theory of genetic contributions, which demonstrates the fundamental relationship of inbreeding and selection, implies that popular sires are the major contributor to high rate of inbreeding.
Evaluating the performance of selection scans to detect selective sweeps in domestic dogs. Florencia Schlamp, Julian van der Made, Rebecca Stambler, Lewis Chesebrough, Adam R Boyko, Philipp W Messer. BioRxiv. October 2015. Quote: "Selective breeding of dogs has resulted in repeated artificial selection on breed-specific morphological phenotypes. A number of quantitative trait loci associated with these phenotypes have been identified in genetic mapping studies. We analyzed the population genomic signatures observed around the causal mutations for 12 of these loci in 25 dog breeds, for which we genotyped 25 individuals in each breed. By measuring the population frequencies of the causal mutations in each breed, we identified those breeds in which specific mutations most likely experienced positive selection. These instances were then used as positive controls for assessing the performance of popular statistics to detect selection from population genomic data. We found that artificial selection during dog domestication has left characteristic signatures in the haplotype and nucleotide polymorphism patterns around selected loci that can be detected in the genotype data from a single population sample. However, the sensitivity and accuracy at which such signatures were detected varied widely between loci, the particular statistic used, and the choice of analysis parameters. We observed examples of both hard and soft selective sweeps and detected strong selective events that removed genetic diversity almost entirely over regions >10 Mbp. Our study demonstrates the power and limitations of selection scans in populations with high levels of linkage disequilibrium due to severe founder effects and recent population bottlenecks."
Serum Biochemical Phenotypes in the Domestic Dog. Yu-Mei Chang, Erin Hadox, Balazs Szladovits, Oliver A. Garden. PlosOne. February 2016. Quote: "The serum or plasma biochemical profile is essential in the diagnosis and monitoring of systemic disease in veterinary medicine, but current reference intervals typically take no account of breed-specific differences. Breed-specific hematological phenotypes have been documented in the domestic dog, but little has been published on serum biochemical phenotypes in this species. Serum biochemical profiles of dogs in which all measurements fell within the existing reference intervals were retrieved from a large veterinary database. Serum biochemical profiles from 3045 dogs were retrieved, of which 1495 had an accompanying normal glucose concentration. Sixty pure breeds plus a mixed breed control group were represented by at least 10 individuals. All analytes, except for sodium, chloride and glucose, showed variation with age. Total protein, globulin, potassium, chloride, creatinine, cholesterol, total bilirubin, ALT, CK, amylase, and lipase varied between sexes. Neutering status significantly impacted all analytes except albumin, sodium, calcium, urea, and glucose. Principal component analysis of serum biochemical data revealed 36 pure breeds with distinctive phenotypes. Furthermore, comparative analysis identified 23 breeds with significant differences from the mixed breed group in all biochemical analytes except urea and glucose. Eighteen breeds were identified by both principal component and comparative analysis. Tentative reference intervals were generated for breeds with a distinctive phenotype identified by comparative analysis and represented by at least 120 individuals [including 174 cavalier King Charles spaniels]. This is the first large-scale analysis of breed-specific serum biochemical phenotypes in the domestic dog and highlights potential genetic components of biochemical traits in this species."
Dog breed affiliation with a forensically validated canine STR set. Burkhard Berger, Cordula Berger, Josephin Heinrich, Harald Niederstätter, Werner Hecht, Andreas Hellmann, Udo Rohleder, Uwe Schleenbecker, Nadja Morf, Ana Freire-Aradas, Dennis McNevin, Christopher Phillips, Walther Parson. Forensic Sci. Int'l: Genetics. November 2-18;37:126-134. Quote: We tested a panel of 13 highly polymorphic canine short tandem repeat (STR) markers for dog breed assignment using 392 dog samples from the 23 most popular breeds in Austria, Germany, and Switzerland. This STR panel had originally been selected for canine identification. The dog breeds sampled in this study featured a population frequency ≥1% and accounted for nearly 57% of the entire pedigree dog population in these three countries. Breed selection was based on a survey comprising records for nearly 1.9 million purebred dogs belonging to more than 500 different breeds. To derive breed membership from STR genotypes, a range of algorithms were used. These methods included discriminant analysis of principal components (DAPC), STRUCTURE, GeneClass2, and the adegenet package for R. STRUCTURE analyses suggested 21 distinct genetic clusters. Differentiation between most breeds was clearly discernable. Fourteen of 23 breeds (61%) exhibited maximum mean cluster membership proportions of more than 0.70 with a highest value of 0.90 found for Cavalier King Charles Spaniels. Dogs of only 6 breeds (26%) failed to consistently show only one major cluster. The DAPC method yielded the best assignment results in all 23 declared breeds with 97.5% assignment success. The frequency-based assignment test also provided a high success rate of 87%. These results indicate the potential viability of dog breed prediction using a well-established and sensitive set of 13 canine STR markers intended for forensic routine use.

CONNECT WITH US